Written by:
German Sastre (Instituto de Tecnologia Quimica, Valencia, Spain)
and
Julian D. Gale (Curtin University of Technology, Perth, Australia)
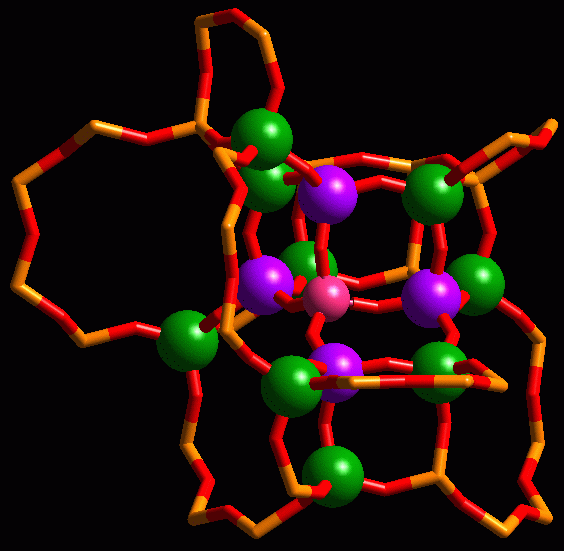
Fragment of the CHA zeotype showing a tetrahedral central atom (pink), and its first (blue) and second (green) T-neighbours.
- 1. Introduction to zeoTsites
- 2. Copyright notice
- 3. Getting zeoTsites
- 4. Input and output files
- 5. Current limitations
- 6. More information
1. Introduction to zeoTsites
The catalytic function in zeolites and zeotypes is strongly determined
by their microporous structure. Microporosity is oftenly viewed only as
the size and shape of the microchannels and microcavities present in
these materials and in this way we are only focusing on the void space.
Such void space is the consecuence of the of the actual directionality
taken by the T-O (T=tetrahedral atom) bonds in the three dimensional
topology of the solid
and forming the void space usually means a deviation of a more stable
topology such as -for example- that present in high density phases
of silicates of equivalent chemical composition. Still less intuitive
is to focus on the connectivity sequence of each particular structure
although some consequences for the catalytic activity may also arise
from it.
Exploring the topology and geometry of three-dimensional four-connected nets is the aim of the zeoTsites Fortran code. For the sake of clarity, the program, apart from calculating every single T-O and T-O-T units, also performs averages. Averages are considered from two viewpoints: T atoms are grouped by their coordination sequence and by their (crystallographic) label. This allows easy comparison between geometry variations related to topology and/or crystallographic site. The program can also be used to analyse geometries of a zeotype in which an isomorphic substitution has been performed and then the response of the framework to the substitution in the different parts of the structure can be analysed.
In the same way, NMR signals coming from different T sites can be classified according to their corresponding average T-O-T angles, or T-T distances, whose values are provided by the code. Any other external perturbation in the zeotype framework by any physical or chemical influence can be analysed by the code if the corresponding geometries are previously provided. In this sense the code can be a useful tool to be utilised after a given computer simulation.
2. Copyright notice
zeoTsites is available free of charge to academic and research institutions and non-commercial establishments only. Copies should be obtained from the authors only and should not be distributed in any form by the user to a third party without the express permission of the authors.
No claim is made that the program is free from errors and no liability will be accepted for any loss or damage that may result. The users are responsible for checking the validity of their results.
3. Getting zeoTsites
A tar file (zeots-x.y.tar, where x.y is the version number) can be obtained
from any of the authors by e-mail.
The unix installation proceeds as follows:
(i) untar the original file zeots-1.2.tar by typing:
'tar xvf zeots-1.2.tar'
(ii) type: 'make'
As to the use of the code:
(i) execute by typing: './zeots'
(ii) you will then be prompted twice as follows:
- "What is the zeotype ?"
you have to provide a name (no more than 20 characters) which
will be appended to the output files
just give the complete name of the input file (.xtl format)
4. Input and output files
zeoTsites is provided with some sample input files (in .xtl format), and it produces 4 output files:
- "Connecty_of_filename"
this contains the coordination sequence of each T-atom in the unit
cell (up to the 13th shell), and it classifies each topologically
distinct atom with a number (in the column labelled "T-type")
then the vertex symbol of each T-atom is printed. For 4-connected
atoms, this consists of 6 symbols corresponding to the rings found
in the: O1-T-O2, O1-T-O3, O1-T-O4, O2-T-O3, O2-T-O4, O3-T-O4
directions. Each index indicates the number of T atoms in the ring,
and the sub-index indicates the number of them. The vertex symbols
are ordered (for 4-connected atoms) according to: ring size,
opposite angles, and ring multiplicity. Opposite angles are defined
(for 4-connected atoms) as: 1-4 and 2-3; 1-3 and 2-4; 1-2 and 3-4.
For connectivities other than 4, the opposite angle criterion is
not taken into account when ordering the vertex symbols.
then it prints the "Geometry analysis:" with the characteristics
of all the T-O-T units in the unit cell (T-type, atom numbers,
T-O-T angle, T-O and T-T distances, and atom labels). Atoms are
renumbered: first all T-atoms, and then all O-atoms. The same
numbering is followed in the "output_filename.xtl" file
then the "Average topological geometry analysis:" is printed which
consists on averages over T-T types and T-types. The number under
the column "Number" indicates the corresponding multiplicity
finally the "Average label geometry analysis:" is printed and these
averages are referred to the label of the T-atoms in the input
file. These averages can be particularly useful if the T and O atoms
are labelled according to their crystallographic positions. Averages
include T-O, T-T, T-O-T, and T averages
- "T1_sites_of_filename"
for each T-atom in the unit cell its T-first-neighbours are
output by indicating: atom number, atom label, cell code
atoms connected to a given atom can be in one of the adjacent
unit cells. the initial cell code is referred to as "0 0 0".
the other cells are indicated by numbers of relative shifts to
the central cell in the directions of the x,y,z axis respectively.
i.e. the contiguous cell at the right would be "1 0 0"
- "T2_sites_of_filename"
for each T-atom in the unit cell its T-second-neighbours are
output by indicating: atom number, atom label, cell code
T1_ and T2_ files can be useful for NMR applications. Complex unit
cells with a given distribution of Si,Al,P atoms can be used as
input files in order to find the neighbourhood of each T-atom
- "output_filename.xtl"
this is a .xtl file with the same input structure which contains
the topological T-type (divided by 1000) in the column corresponding
to the charges. This is useful for visual identification of the
T-types when using a visualisation software such as Cerius2
5. Current limitations
(i) T-T connectivity is calculated through distance checks and confirmed
by label check. Non connected T-T atoms in the range indicated by the
parameters (adist1,adist2) will be identified as connected and the
program will fail
(ii) cations must be cleared from input file as they do not contribute to
the framework connectivity and they are not taken into account in
the geometry analysis. Otherwise some cations may be interpreted as
bonded to some of the framework atoms thus flawing the corresponding
connectivity and spoiling the geometry analysis
6. More information
See the following paper:
Title: ZeoTsites: a code for topological and crystallographic tetrahedral
sites analysis in zeolites and zeotypes
Authors: G. Sastre, J.D. Gale
Journal: Microporous and Mesoporous Materials
Year: 2001
Volume: 43
Issue: 1
Pages: 27-40